Introduction to Spatial Data
GEO 200CN - Quantitative Geography
Professor Noli Brazil
April 29, 2024
This week you will learn how to process and map spatial data in R. The two major packages for processing spatial data in R are sf and terra. This guide will provide an introduction to these packages. To motivate the use of these packages, the bulk of the guide will focus on how to map data, which will help you answer the mapping questions in this week’s assignment.
Installing and loading packages
Install sf and terra if you have not already done so.
install.packages(c("sf", "terra"))We will also be using the packages tmap, RColorBrewer, maptiles, and cartogram in this lab.
install.packages(c("tmap", "RColorBrewer", "maptiles", "cartogram"))Load all of these packages using library(). We’ll also
be using the package tidyverse, so load it up as well.
Remember, you always need to use library() at the top of
your R Markdown files.
library(sf)
library(terra)
library(tmap)
library(tidyverse)
library(RColorBrewer)
library(maptiles)
library(cartogram)Spatial data
Spatial phenomena can generally be thought of as either discrete objects with clear boundaries or as a continuous phenomenon that can be observed everywhere, but does not have natural boundaries. Discrete spatial objects may refer to a river, road, country, town, or a research site. Examples of continuous phenomena, or “spatial fields”, include elevation, temperature, and air quality.
There are two general spatial data object types: Vector and Raster. Areal data are typically usually represented with a vector data structure. Such data consists of a description of the “geometry” or “shape” of the objects, and normally also includes additional variables. For example, a vector data set may describe the borders of the countries of the world (geometry), and also store their names and the size of their population; or the geometry of the roads in an area, as well as their type and names. These additional variables are often referred to as “attributes”. Continuous spatial data (fields) are usually represented with a raster data structure. Raster datasets are simply an array of pixels/cells organized into rows and columns (or a grid) where each cell contains a value representing information, such as temperature, soil type, land use, water level. Raster maps usually represent continuous phenomena such as elevation, temperature, population density or spectral data. Discrete features such as soil or land-cover classes can also be represented in the raster data model. Rasters are also aerial photographs, imagery from satellites, Google street view images.
We discuss these vector and raster data types, and how they are processed in R, in turn.
Vector data
The main vector data types are points, lines and polygons. In all cases, the geometry of these data structures consists of sets of coordinate pairs (x, y). Points are the simplest case. Each point has one coordinate pair, and n associated variables. For example, a point might represent a place where a rat was trapped, and the attributes could include the date it was captured, the person who captured it, the species size and sex, and information about the habitat. It is also possible to combine several points into a multi-point structure, with a single attribute record. For example, all the coffee shops in a town could be considered as a single geometry.
The geometry of lines is a just a little bit more complex. First note that in this context, the term ‘line’ refers to a set of one or more polylines (connected series of line segments). For example, in spatial analysis, a river and all its tributaries could be considered as a single ‘line’ (but they could also also be several lines, perhaps one for each tributary river). Lines are represented as ordered sets of coordinates (nodes). The actual line segments can be computed (and drawn on a map) by connecting the points. Thus, the representation of a line is very similar to that of a multi-point structure. The main difference is that the ordering of the points is important, because we need to know which points should be connected. A network (e.g. a road or river network), or spatial graph, is a special type of lines geometry where there is additional information about things like flow, connectivity, direction, and distance.
A polygon refers to a set of closed polylines. The geometry is very similar to that of lines, but to close a polygon the last coordinate pair coincides with the first pair. Multiple polygons can be considered as a single geometry. For example, the Philippines consists of many islands. Each island can be represented by a single polygon, but together they can be represented as a single (multi-) polygon representing the entire country.
Two R packages that handle vector data are terra and sf. Although there are overlaps between the two packages, and you can convert vector objects represented in one package into the other, the packages handle vector data in unique ways. We cover each package below.
terra package
The terra package is primarily for creating, reading, manipulating, and writing raster data. However, it also handles vector data. In general, the package defines a set of classes to represent spatial data. These classes start with the name Spat. For vector data, the relevant class is SpatVector. These classes represent geometries as well as attributes (variables) describing the geometries. The main reason for defining classes is to create a standard representation of a particular data type to make it easier to write functions (also known as ‘methods’) for them.
To demonstrate how vector data are generally depicted in
terra, let’s bring in the polygon shapefile
lux, which is a shapefile of Luxembourg and is a
part of the terra package. The shapefile
is the most commonly used file format for vector data. Use the following
code to to get the full path name of the file’s location. We need to do
this as the location of this file depends on where the
terra package is installed. You should not use the
system.file() function for your own files. In other words,
don’t dwell too much on what this function is doing. It only serves for
creating examples with data that ships with R.
filename <- system.file("ex/lux.shp", package="terra")
filename## [1] "/Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/library/terra/ex/lux.shp"Now we have the filename and its path that we need to use. To bring
it into R, use the function vect(), which is a part of the
terra package.
lux <- vect(filename)We find that it is a SpatVector object.
class(lux)## [1] "SpatVector"
## attr(,"package")
## [1] "terra"We can use the base plot() function to make a map of
Luxembourg’s regions.
plot(lux)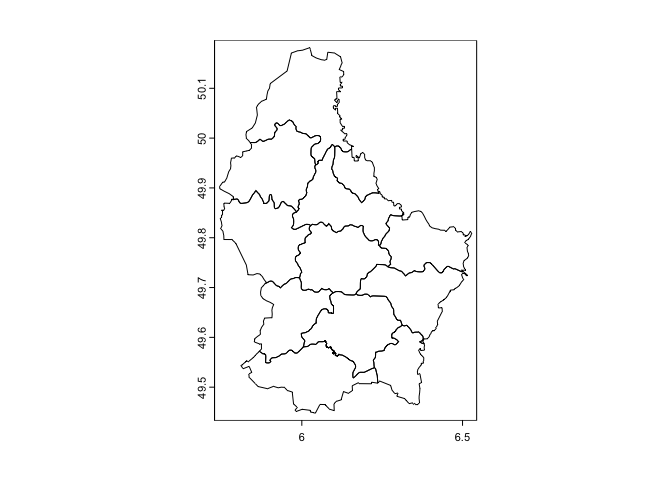
We mapped the polygons above, but we can also map one of the attributes. Here we show a choropleth map of the area of each region AREA.
plot(lux, 'AREA')
Or plot a categorical variable such as the name of each region
plot(lux, 'NAME_2')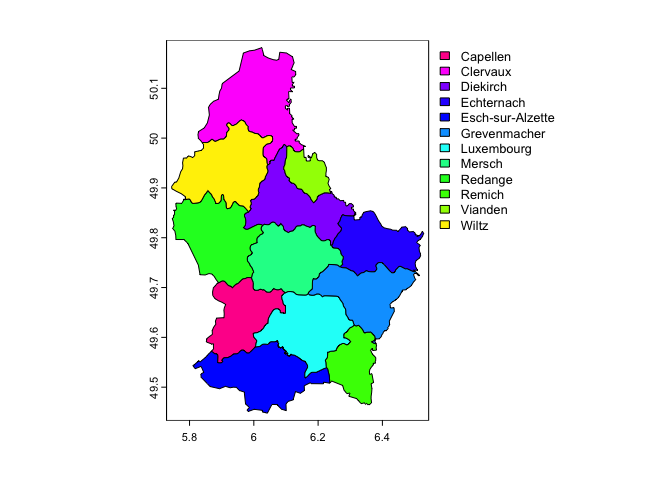
We can manipulate the SpatVector like we would a regular data frame. Here we’ll use base R to manipulate. To extract the attributes (data.frame) from a Spatial object, use:
d <- as.data.frame(lux)
head(d)## ID_1 NAME_1 ID_2 NAME_2 AREA POP
## 1 1 Diekirch 1 Clervaux 312 18081
## 2 1 Diekirch 2 Diekirch 218 32543
## 3 1 Diekirch 3 Redange 259 18664
## 4 1 Diekirch 4 Vianden 76 5163
## 5 1 Diekirch 5 Wiltz 263 16735
## 6 2 Grevenmacher 6 Echternach 188 18899You can extract a variable using the $ convention.
lux$NAME_2## [1] "Clervaux" "Diekirch" "Redange" "Vianden"
## [5] "Wiltz" "Echternach" "Remich" "Grevenmacher"
## [9] "Capellen" "Esch-sur-Alzette" "Luxembourg" "Mersch"Can we use tidyverse functions on SpatVectors?
lux %>%
select(NAME_2)## Error in UseMethod("select"): no applicable method for 'select' applied to an object of class "SpatVector"Oh oh. terra is not tidy friendly. Hence, why base R is still important to understand, as some important packages have not integrated the tidy workflow (although, see the package tidyterra as an attempt to bridge this integration gap).
Create a new variable, for example a new variable containing AREA multiplied by 100
lux$AREA_100 <- lux$AREA*100Now, sub-setting by variable.
lux[, 'NAME_2']## class : SpatVector
## geometry : polygons
## dimensions : 12, 1 (geometries, attributes)
## extent : 5.74414, 6.528252, 49.44781, 50.18162 (xmin, xmax, ymin, ymax)
## source : lux.shp
## coord. ref. : lon/lat WGS 84 (EPSG:4326)
## names : NAME_2
## type : <chr>
## values : Clervaux
## Diekirch
## RedangeNote how this code is different from the code above it. Above
lux$NAME_2 returns a vector of values is returned. With the
approach directly above you get a new SpatVector with only one
variable.
Selecting rows (records).
g <- lux[lux$NAME_1 == 'Grevenmacher',]
plot(g)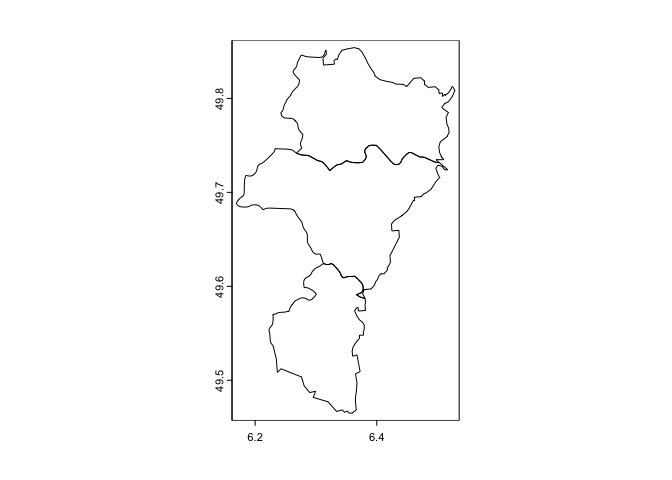
There are other terra spatial data functions you can use to manipulate the polygons. Since we won’t be focusing on the terra package for handling vector data in this class, you can learn more about these functions on your own here.
Although useful when working with vector data within the terra universe, it is not the current standard package for processing vector data in R. This designation belongs to the sf package, which was created in 2016. In contrast to the terra package, which uses a rather complex data structure, sf is much easier to use and fits in very naturally with the tidyverse. Which means we can use most of the functions we went through when we learned about the tidyverse package to clean and process sf spatial objects. This includes mapping vector data using ggplot, which we will cover later.
sf package
The sf package is the major package for handling vector data in R. It conceives of spatial objects as simple features. What is a feature? A feature is thought of as a thing, or an object in the real world, such as a building or a tree. A wetland preserve can be a feature. As can a city and a neighborhood. Features have a geometry describing where on Earth the features are located, and they have attributes, which describe other properties. The main application of simple feature geometries is to describe two-dimensional geometries by points, lines, or polygons.
Reading spatial data
The function for reading in spatial data into R as an
sf object is st_read(). I zipped up and
uploaded onto GitHub a folder containing files for this lab. Set your
working directory to an appropriate folder and use the following code to
download and unzip the file.
#insert the pathway to the folder you want your data stored into
setwd("insert your pathway here")
#downloads file into your working directory
download.file(url = "https://raw.githubusercontent.com/geo200cn/data/master/spatialsflab.zip", destfile = "spatialsflab.zip")
#unzips the zipped file
unzip(zipfile = "spatialsflab.zip")You should see the shapefiles saccityinc.shp,
Rivers.shp, Parks.shp, and EV_Chargers.shp in
the folder you specified in setwd(). These files contain
Sacramento city Census
tract polygons, Sacramento river polylines, Sacramento parks
polygons, and Sacramento
Electric Vehicle Charging Locations points, respectively. You should
also see a csv file sacrace.csv, which contains Sacramento city
Census tract race/ethnic population counts. First, bring in the
shapefiles using the function st_read().
saccitytracts <- st_read("saccityinc.shp", stringsAsFactors = FALSE)
parks <- st_read("Parks.shp", stringsAsFactors = FALSE)
rivers <- st_read("Rivers.shp", stringsAsFactors = FALSE)
evcharge <- st_read("EV_Chargers.shp", stringsAsFactors = FALSE)The argument stringsAsFactors = FALSE tells R to keep
any variables that look like a character as a character and not a
factor.
Using class() we find that they are sf
objects
class(saccitytracts)## [1] "sf" "data.frame"Look at saccitytracts
saccitytracts## Simple feature collection with 122 features and 2 fields
## Geometry type: MULTIPOLYGON
## Dimension: XY
## Bounding box: xmin: -121.5601 ymin: 38.43789 xmax: -121.3627 ymax: 38.6856
## Geodetic CRS: NAD83
## First 10 features:
## GEOID medinc geometry
## 1 6067007104 High MULTIPOLYGON (((-121.5076 3...
## 2 6067000400 High MULTIPOLYGON (((-121.4748 3...
## 3 6067006900 Low MULTIPOLYGON (((-121.4729 3...
## 4 6067003102 Medium MULTIPOLYGON (((-121.4457 3...
## 5 6067006400 Low MULTIPOLYGON (((-121.4291 3...
## 6 6067001101 Low MULTIPOLYGON (((-121.4989 3...
## 7 6067009633 Medium MULTIPOLYGON (((-121.4476 3...
## 8 6067001900 Medium MULTIPOLYGON (((-121.4824 3...
## 9 6067009614 High MULTIPOLYGON (((-121.4447 3...
## 10 6067004906 Medium MULTIPOLYGON (((-121.4531 3...The output gives us pertinent information regarding the spatial file, including the type (MULTIPOLYGON), the extent or bounding box, the coordinate reference system, and the 3 variables in the attribute dataset. Most important is the geometry column, which tells you that the object is in spatial format. Click on saccitytracts in your environment window. You’ll find that a window pops up in the top left portion of your interface. Unlike with SpatVector objects, an sf object looks and feels like a regular data frame. The only difference is there is a geometry column. This column “spatializes” the dataset, or lets R know where each feature is geographically located on the Earth’s surface. Check out the other datasets we brought in and determine how they differ from one another.
You can bring in other types of spatial data files other than shapefiles. See a list of these file types here.
Next bring in sacrace.csv which contains Hispanic
(hisp), non-Hispanic Asian (nhasn), non-Hispanic Black
(nhblk), non-Hispanic white (nhwhite) and total
population (tpopr) for tracts in Sacramento city. Use the
function read_csv().
sacrace <- read_csv("sacrace.csv")Data manipulation
sf objects are data frames. This means that they are
tidy friendly. You can use many of the functions we learned previously to manipulate
sf objects, and this includes our new best friend the
pipe %>% operator. For example, let’s do the following
tasks on saccitytracts.
- Merge in the race/ethnicity and total population variables from
sacrace into saccitytracts using
left_join(). The common ID is GEOID - Create the variable pwh which is the percent of the tract
population that is non-Hispanic white using
mutate() - Keep just the variables GEOID, medinc, and
pwh using the function
select()
We do this in one line of continuous code using the pipe operator
%>%
saccitytracts <- saccitytracts %>%
left_join(sacrace, by = "GEOID") %>%
mutate(pwh = nhwhite/tpopr) %>%
select(GEOID, medinc, pwh)You can also keep certain polygons (i.e. subset rows). For example, keep the tracts that are majority non-white
mwhite <- saccitytracts %>%
filter(pwh < 0.5)
#number of neighborhoods in Sacramento that are majority non white
nrow(mwhite)## [1] 94The main takeaway: sf objects are tidy friendly, so you can use many of the tidy functions on these objects to manipulate your data set.
While tidyverse offers a set of functions for data
manipulation, the sf package offers a suite of
functions unique to manipulating spatial data. Spatial data manipulation
involves cleaning or altering your data set based on the geographic
location of features. Most of these functions start out with the prefix
st_.
Common functions to manipulate sf objects include the following:
st_read()reads a sf object,st_write()writes a sf object,st_crs()gets or sets a new coordinate reference system (CRS),st_transform()transforms data to a new CRS,st_intersection()intersects sf objects,st_union()combines several sf objects into one,st_simplify()simplifies a sf object,st_coordinates()retrieves coordinates of a sf object,st_as_sf()converts a foreign object to a sf object.
To see all of the functions, type in
methods(class = "sf")We won’t go through these functions as the list is quite extensive. You can learn about them in Geocomputation in R’s excellent guide. The focus of this lab is to learn how to map sf objects, which we’ll get to later in this guide.
Raster data
Raster data is commonly used to represent spatially continuous phenomena such as elevation. A raster divides the world into a grid of equally sized rectangles (referred to as cells or, in the context of satellite remote sensing, pixels) that all have one or more values (or missing values) for the variables of interest. A raster cell value should normally represent the average (or majority) value for the area it covers. However, in some cases the values are actually estimates for the center of the cell (in essence becoming a regular set of points with an attribute).
In contrast to vector data, in raster data the geometry is not explicitly stored as coordinates. It is implicitly set by knowing the spatial extent and the number of rows and columns in which the area is divided. From the extent and number of rows and columns, the size of the raster cells (spatial resolution) can be computed. While raster cells can be thought of as a set of regular polygons, it would be very inefficient to represent the data that way as coordinates for each cell would have to be stored explicitly. It would also dramatically increase processing speed in most cases.
The main package for handling raster data in R is terra. The package replaces the package raster. The change was made because terra offers faster processing speeds and more flexible functions. The package provides, among other things, general raster data manipulation functions that can easily be used to develop more specific functions. For example, there are functions to read a chunk of raster values from a file or to convert cell numbers to coordinates and back. The package also implements raster algebra and many other functions for raster data manipulation.
The package is built around a number of “classes” of which the SpatVector we already met earlier. The raster class is SpatRaster, which we introduce next.
SpatRaster
A SpatRaster represents multi-layer (multi-variable) raster data. A SpatRaster always stores a number of fundamental parameters describing its geometry. These include the number of columns and rows, the spatial extent, and the Coordinate Reference System. In addition, a SpatRaster can store information about the file in which the raster cell values are stored. Or, if there is no such a file, a SpatRaster can hold the cell values in memory.
A SpatRaster can easily be created from scratch using the
function rast(). The default settings will create a global
raster data structure with a longitude/latitude coordinate reference
system and 1 by 1 degree cells. You can change these settings by
providing additional arguments such as xmin, nrow,
ncol, and/or crs, to the function. You can also change
these parameters after creating the object. If you set the projection,
this is only to properly define it, not to change it. To transform a
SpatRaster to another coordinate reference system (projection)
you can use the function warp(). But note that in most
cases where real data is analyzed, these objects are created from a
file.
Let’s start by creating a SpatRaster from scratch using the
rast() function.
r <- rast(ncol=10, nrow=10, xmin=-150, xmax=-80, ymin=20, ymax=60)
r## class : SpatRaster
## dimensions : 10, 10, 1 (nrow, ncol, nlyr)
## resolution : 7, 4 (x, y)
## extent : -150, -80, 20, 60 (xmin, xmax, ymin, ymax)
## coord. ref. : lon/lat WGS 84SpatRaster r only has the geometry of a raster data
set. That is, it knows about its location, resolution, etc., but there
are no values associated with it. We check this using the function
hasValues().
hasValues(r)## [1] FALSELet’s assign some values. In this case we assign a vector of random
numbers with a length that is equal to the number of raster cells in
r using the function runif().
values(r) <- runif(ncell(r))
r## class : SpatRaster
## dimensions : 10, 10, 1 (nrow, ncol, nlyr)
## resolution : 7, 4 (x, y)
## extent : -150, -80, 20, 60 (xmin, xmax, ymin, ymax)
## coord. ref. : lon/lat WGS 84
## source(s) : memory
## name : lyr.1
## min value : 0.02077911
## max value : 0.99080994You could also assign cell numbers (in this case overwriting the previous values)
values(r) <- 1:ncell(r)
r## class : SpatRaster
## dimensions : 10, 10, 1 (nrow, ncol, nlyr)
## resolution : 7, 4 (x, y)
## extent : -150, -80, 20, 60 (xmin, xmax, ymin, ymax)
## coord. ref. : lon/lat WGS 84
## source(s) : memory
## name : lyr.1
## min value : 1
## max value : 100We can plot this object using base plot().
plot(r)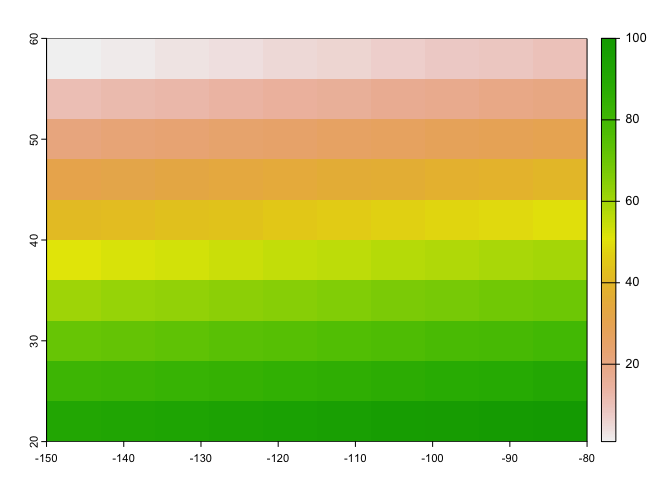
You can create a multi-layer (or multi band) object using the
c method.
r2 <- r * r
r3 <- sqrt(r)
s <- c(r, r2, r3)
s## class : SpatRaster
## dimensions : 10, 10, 3 (nrow, ncol, nlyr)
## resolution : 7, 4 (x, y)
## extent : -150, -80, 20, 60 (xmin, xmax, ymin, ymax)
## coord. ref. : lon/lat WGS 84
## source(s) : memory
## names : lyr.1, lyr.1, lyr.1
## min values : 1, 1, 1
## max values : 100, 10000, 10Think of r, r2, and r3 as representing different cell values for the same grid (e.g., temperature across three points in time). And plot this
plot(s)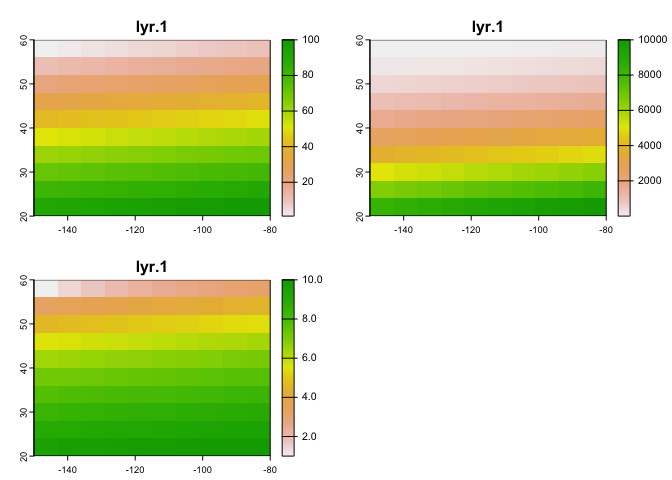
Reading in raster data
In most applications you will bring in a raster dataset directly from a file. The raster package can use raster files in several formats, including some ‘natively’ supported formats and other formats via the rgdal package. Supported formats for reading include GeoTIFF, ESRI, ENVI, and ERDAS. Most formats supported for reading can also be written to.
A notable feature of the terra package is that it can work with raster datasets that are stored on disk and are too large to be loaded into memory (RAM). The package can work with large files because the objects it creates from these files only contain information about the structure of the data, such as the number of rows and columns, the spatial extent, and the filename, but it does not attempt to read all the cell values in memory. In computations with these objects, data is processed in chunks. If no output filename is specified to a function, and the output raster is too large to keep in memory, the results are written to a temporary file.
Here is an example using the elev dataset (taken from the
terra package), using a file in the native
‘raster-file’ format. Use the following code to bring in the data. The
file provides elevation values for Luxembourg. Note do not use this
system.file construction for your own files. Just type the
file name as you would do for any other file, but don’t forget to use
forward slashes as path separators.
filename <- system.file("ex/elev.tif", package="terra")
basename(filename)## [1] "elev.tif"Now bring in the file using the function rast()
elev <- rast(filename)
elev## class : SpatRaster
## dimensions : 90, 95, 1 (nrow, ncol, nlyr)
## resolution : 0.008333333, 0.008333333 (x, y)
## extent : 5.741667, 6.533333, 49.44167, 50.19167 (xmin, xmax, ymin, ymax)
## coord. ref. : lon/lat WGS 84 (EPSG:4326)
## source : elev.tif
## name : elevation
## min value : 141
## max value : 547We see that this data is a SpatRaster. Remember that raster
data are basically grid data, and the grid we have contains 90 rows, 95
columns, 8550 (90 x 95) cells. There’s also just 1 layer (elevation).
Let’s plot what we got (elevation values) using plot().
plot(elev)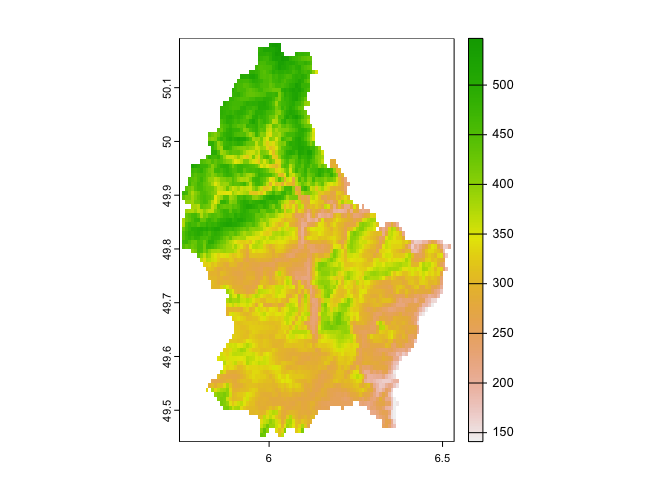
Vector to raster conversion
Rather than bringing in a raster dataset, you often have to convert a vector object, typically polygons or points, into a raster. Polygon to raster conversion is typically done to create a SpatRaster that can act as a mask, i.e. to set to NA a set of cells of a raster object, or to summarize values on a raster by zone. For example a country polygon is transferred to a raster that is then used to set all the cells outside that country to NA; whereas polygons representing administrative regions such as states can be transferred to a raster to summarize raster values by region.
Let’s turn the Luxembourg polygon lux we used earlier into a
raster. In order to change it into a raster object, use the function
rast(), and designate the raster grid (number of
cells).
r <- rast(lux, ncols=75, nrows=100 )The size of each pixel or cell in the raster grid is defined by the
number of columns ncols and rows nrows. You
can also defines the resolution using the res argument.
Plor r
plot(r)
You should get a blank map. Object r only has the skeleton
of a raster data set. That is, the object r created only
consists of the raster geometry, that is, we have defined the number of
rows and columns, and where the raster is located in geographic space,
but there are no cell-values associated with it. The function
rasterize()transfers values associated with ‘object’ type
spatial data (points, lines, polygons) to raster cells. Let’s transfer
the area AREA from lux to their associated raster cell
in r.
vr <- rasterize(lux, r, 'AREA')
plot(vr)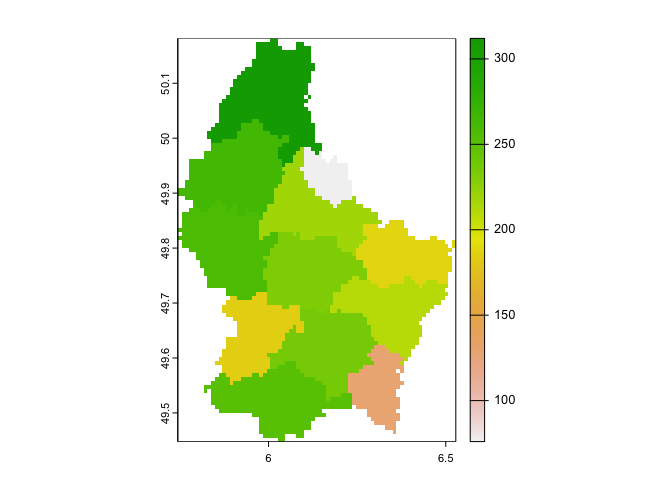
Notice the difference between how this map “looks” compared to the vector data mapped earlier. Why is there a difference?
Point to raster conversion is often done with the purpose to analyze
the point data. For example to count the number of distinct species
(represented by point observations) that occur in each raster cell.
rasterize() takes a SpatRaster object to set the
spatial extent and resolution, and a function to determine how to
summarize the points (or an attribute of each point) by cell.
Similar to vector data, you can manipulate raster data, such assign values to cells, add two rasters, and so on. We won’t go into too much of this until later in the class when we go through spatial interpolation, but you can learn more about these functions here.
Mapping
Now that you’ve learned the basics of bringing in and manipulating
spatial data, let’s go through how we can map it. We already did some
mapping above, but let’s go through the set of map types discussed in
OSU Ch. 3. For vector data, we’ll focus entirely on mapping
sf spatial objects. There are two ways of mapping
sf objects: ggplot() from the
ggplot2 package and tm_shape() from the
tmap package.
ggplot
Because sf is tidy friendly, it is no surprise we
can use the tidyverse plotting function ggplot() to make
maps. We already received an introduction to ggplot() a few labs ago. Recall its
basic structure:
ggplot(data = <DATA>) +
<GEOM_FUNCTION>(mapping = aes(x, y)) +
<OPTIONS>()In mapping, geom_sf() is
<GEOM_FUNCTION>(). Unlike with functions like
geom_histogram() and geom_boxplot(), we don’t
specify an x and y axis. Instead you use fill if you want to map a
variable or color to just map boundaries.
Let’s use ggplot() to make a choropleth map. Choropleth
maps are discussed on page 73-76 in OSU. We need to specify a numeric
variable in the fill = argument within
geom_sf(). Here we map percent non-Hispanic white
pwh.
ggplot(data = saccitytracts) +
geom_sf(aes(fill = pwh))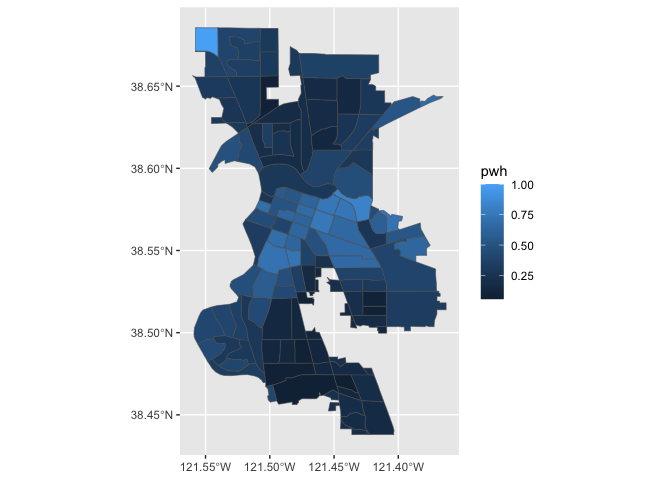
We can also specify a title (as well as subtitles and captions) using
the labs() function.
ggplot(data = saccitytracts) +
geom_sf(aes(fill = pwh)) +
labs(title = "Percent white Sacramento City Tracts") 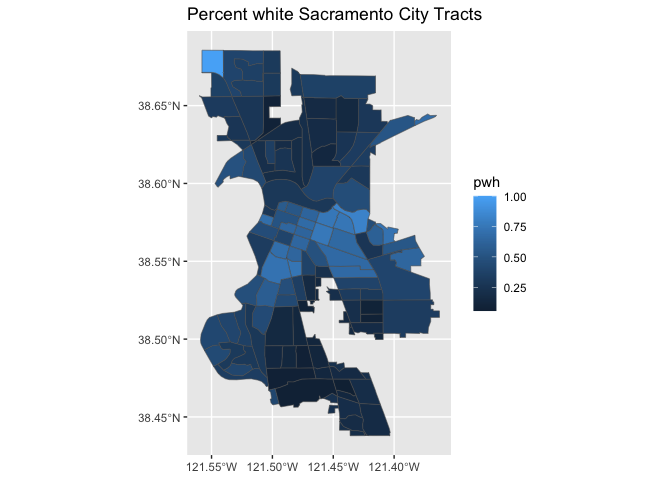
We can make further layout adjustments to the map. Don’t like a blue
scale on the legend? You can change it using the
scale_file_gradient() function. Let’s change it to a white
to red gradient. We can also eliminate the gray tract border colors to
make the fill color distinction clearer. We do this by specifying
color = NA inside geom_sf(). We can also get
rid of the gray background by specifying a basic black and white theme
using theme_bw().
ggplot(data = saccitytracts) +
geom_sf(aes(fill = pwh), color = NA) +
scale_fill_gradient(low= "white", high = "red", na.value ="gray") +
labs(title = "Percent white Sacramento City Tracts",
caption = "Source: American Community Survey") +
theme_bw()
I also added a caption indicating the source of the data using the
captions = parameter within labs().
You also use geom_sf() for mapping points (an example of
a pin map as described in OSU page 66). Color them black using
fill = "black". Let’s map the location of EV chargers in
Sacramento.
ggplot(data = evcharge) +
geom_sf(fill = "black") +
labs(title = "EV Charge Stations Sacramento City Tracts",
caption = "Source: City of Sacramento") +
theme_bw()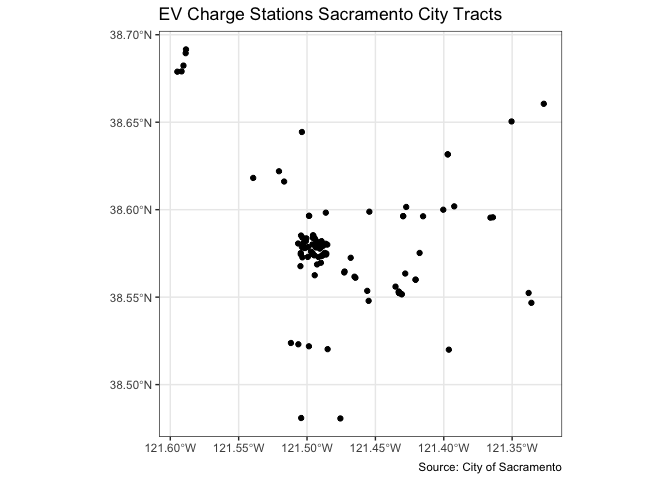
You can overlay the points over Sacramento tracts to give the
locations some perspective. Here, you add two geom_sf() for
the tracts and the EV charge stations.
ggplot() +
geom_sf(data = saccitytracts) +
geom_sf(data = evcharge, fill = "black") +
labs(title = "EV Charge Stations Sacramento City Tracts",
caption = "Source: City of Sacramento") +
theme_bw()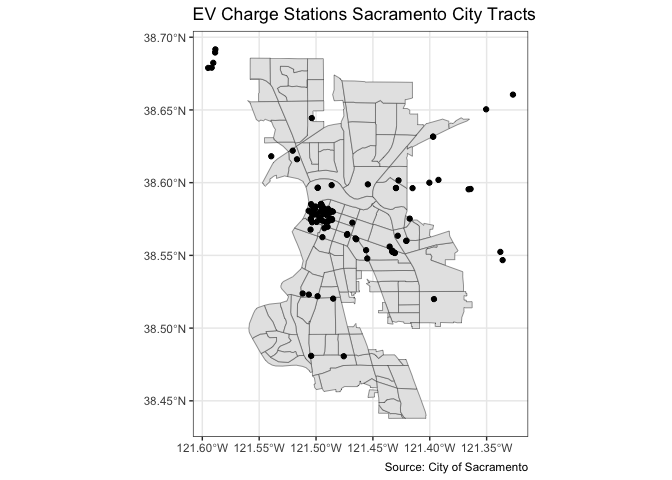
Note that data = moves out of ggplot() and
into geom_sf() because we are mapping more than one spatial
feature.
What about a map of polylines? Here is a map of rivers intersecting with Sacramento City.
ggplot() +
geom_sf(data = rivers, col = "blue") +
labs(title = "Sacramento Rivers",
caption = "Source: Sacramento County") +
theme_bw()
We can use ggplot() to also map raster data. Here, we
plot the raster data set elev we brought earlier, which
represents elevation in Luxembourg. To create a map of raster data using
ggplot, we need to transform the data to a data frame
and pass it to geom_raster(). Let’s also plot the
Luxembourg border by creating an sf object using
st_as_sf().
# Transform data to sf object
luxelev <- st_as_sf(as.data.frame(elev, xy = TRUE), coords = c("x", "y"))
# Assign CRS
st_crs(luxelev) <- 4326
# Plot
ggplot(luxelev) +
geom_sf() +
geom_raster(data = as.data.frame(elev, xy = TRUE),
aes(x = x, y = y, fill = elevation))
Within aes() you specify the x and y coordinates of the
grid points.
tmap
Whether one uses the tmap or ggplot is a matter of taste, but I find that tmap has some benefits, so let’s focus on its mapping functions next.
tmap uses the same layered logic as ggplot. The initial command istm_shape(),
which specifies the geography to which the mapping is applied. This is
followed by a number of tm_* options that select the type of map and
several optional customizations. Check the full list of tm_
elements here.
Choropleth map
Let’s make a choropleth map of percent white in Sacramento.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile")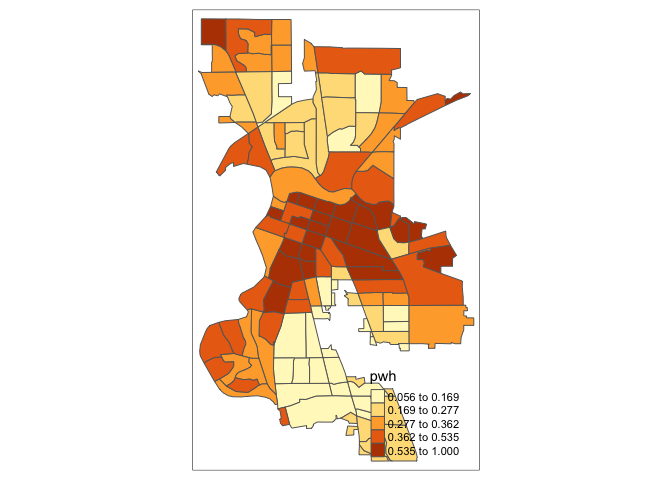
You first put the dataset saccitytracts inside
tm_shape(). Because you are plotting polygons, you use
tm_polygons() next. If you are plotting points, you will
use tm_dots(). If you are plotting lines, you will use
tm_lines(). The argument col = "pwh" tells R
to shade the tracts by the variable pwh. The argument
style = "quantile" tells R to break up the shading into
quintiles, or equal groups of 5. I find that this is where
tmap offers a distinct advantage over
ggplot in that users have greater control over the
legend and bin breaks. tmap allows users to specify
algorithms to automatically create breaks with the style
argument. OSU discusses the importance of breaks and classifications on
page 75. You can also change the number of breaks by setting
n=. The default is n=5. Rather than quintiles,
you can show quartiles using n=4
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile", n=4)
Check out this breakdown of the available classification styles in tmap.
The tm_polygons() command is a wrapper around two other
functions, tm_fill() and tm_borders().
tm_fill() controls the contents of the polygons (color,
classification, etc.), while tm_borders() does the same for
the polygon outlines.
For example, using the same shape (but no variable), we obtain the
outlines of the neighborhoods from the tm_borders()
command.
tm_shape(saccitytracts) +
tm_borders()
Similarly, we obtain a choropleth map without the polygon outlines
when we just use the tm_fill() command.
tm_shape(saccitytracts) +
tm_fill("pwh")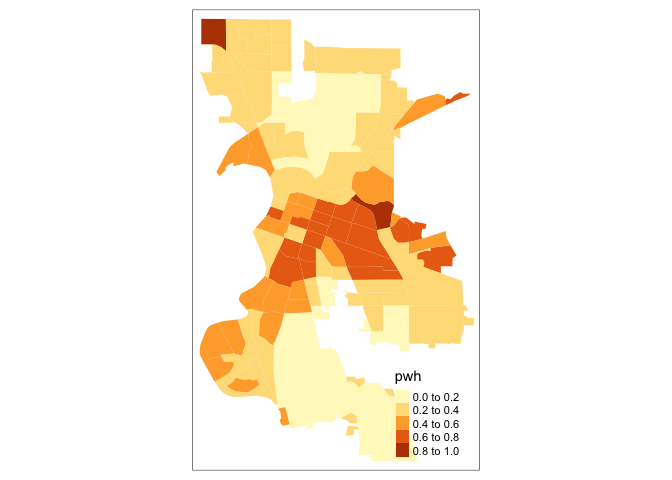
When we combine the two commands, we obtain the same map as with
tm_polygons() (this illustrates how in R one can often
obtain the same result in a number of different ways). Try this on your
own.
You can overlay multiple features on one map. For example, we can add
park polygons on top of city tracts, providing a visual association
between parks and percent white. You will need to add two
tm_shape() functions each for saccitytracts and
parks.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile", n=4) +
tm_shape(parks) +
tm_polygons(col = "green")
Color scheme
The argument palette = defines the color ranges
associated with the bins and determined by the style
arguments. Several built-in palettes are contained in
tmap. For example, using palette = "Reds"
would yield the following map for our example.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds") 
Under the hood, “Reds” refers to one of the color
schemes supported by the RColorBrewer package (see
below).
In addition to the built-in palettes, customized color ranges can be
created by specifying a vector with the desired colors as anchors. This
will create a spectrum of colors in the map that range between the
colors specified in the vector. For instance, if we used
c(“red”, “blue”), the color spectrum would move from red to
purple, then to blue, with in between shades. In our example:
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = c("red","blue")) 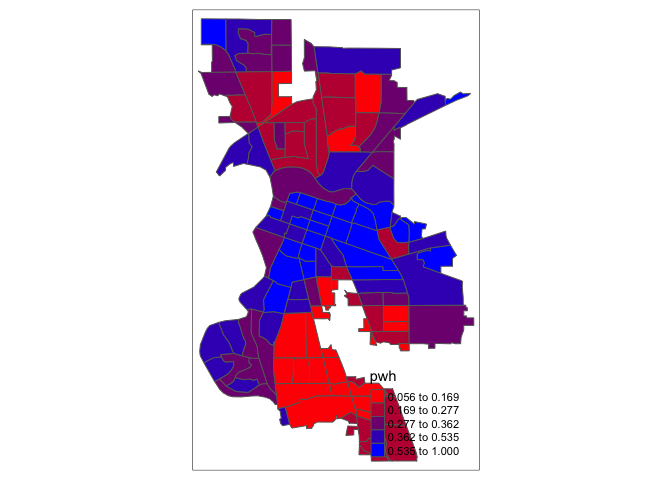
Not exactly a pretty picture. In order to capture a diverging scale, we insert “white” in between red and blue.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = c("red","white", "blue")) 
A preferred approach to select a color palette is to chose one of the schemes contained in the RColorBrewer package. These are based on the research of cartographer Cynthia Brewer (see the colorbrewer2 web site for details). ColorBrewer makes a distinction between sequential scales (for a scale that goes from low to high), diverging scales (to highlight how values differ from a central tendency), and qualitative scales (for categorical variables). For each scale, a series of single hue and multi-hue scales are suggested. In the RColorBrewer package, these are referred to by a name (e.g., the “Reds” palette we used above is an example). The full list is contained in the RColorBrewer documentation.
There are two very useful commands in this package. One sets a color palette by specifying its name and the number of desired categories. The result is a character vector with the hex codes of the corresponding colors.
For example, we select a sequential color scheme going from blue to
green, as BuGn, by means of the command
brewer.pal, with the number of categories (6) and the
scheme as arguments. The resulting vector contains the HEX codes for the
colors.
brewer.pal(6,"BuGn")## [1] "#EDF8FB" "#CCECE6" "#99D8C9" "#66C2A4" "#2CA25F" "#006D2C"Using this palette in our map yields the following result.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette="BuGn") 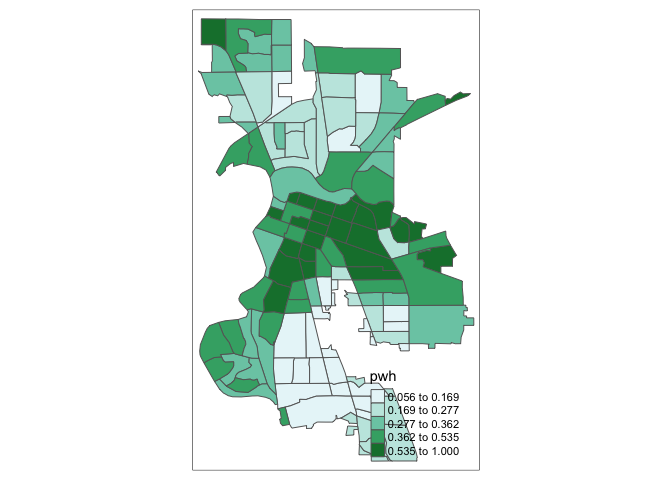
The command display.brewer.pal() allows us to explore
different color schemes before applying them to a map. For example:
display.brewer.pal(6,"BuGn")
Legend
There are many options to change the formatting of the legend. The
automatic title for the legend is not that attractive, since it is
simply the variable name. This can be customized by setting the
title argument.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") 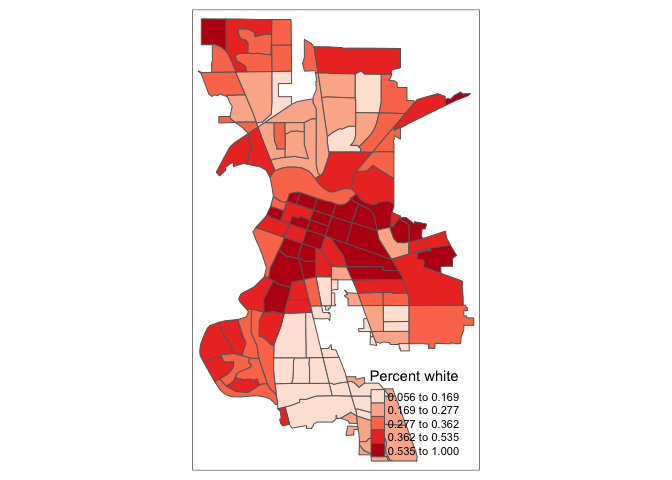
Another important aspect of the legend is its positioning. This is
handled through the tm_layout() function. This function has
a vast number of options, as detailed in the documentation.
There are also specialized subsets of layout functions, focused on
specific aspects of the map, such as tm_legend(),
tm_style() and tm_format(). We illustrate the
positioning of the legend.
The default is to position the legend inside the map. Often, this
default solution is appropriate, but sometimes further control is
needed. The legend.position argument to the
tm_layout function moves the legend around the map, and it
takes a vector of two string variables that determine both the
horizontal position (“left”, “right”, or “center”) and the vertical
position (“top”, “bottom”, or “center”).
For example, if we would want to move the legend to the upper-right position, we would use the following set of commands.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_layout(legend.position = c("right", "top"))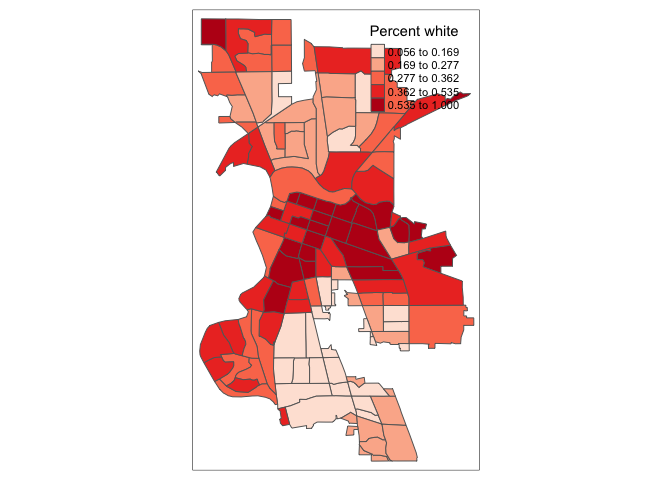
There is also the option to position the legend outside the frame of
the map. This is accomplished by setting legend.outside to
TRUE (the default is FALSE), and optionally also specify its position by
means of legend.outside.position(). The latter can take the
values “top”, “bottom”, “right”, and “left”.
For example, to position the legend outside and on the right, would be accomplished by the following commands.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_layout(legend.outside = TRUE, legend.outside.position = "right")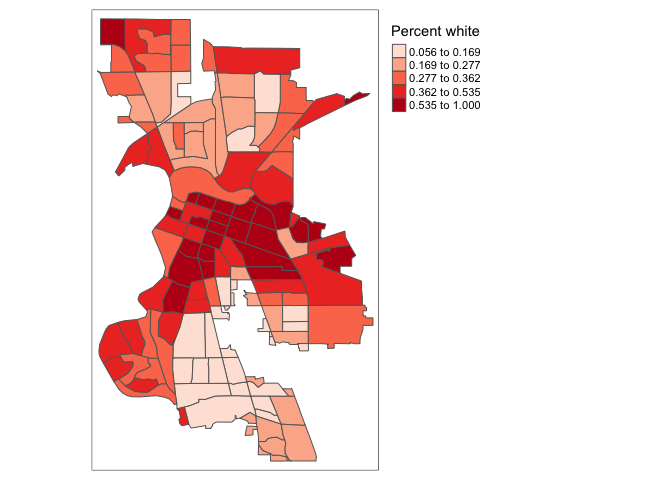
We can also customize the size of the legend, its alignment, font, etc. We refer to the documentation for specifics.
Basemap
For vector data, it’s often nice to display over a basemap by accessing raster tiles that are served on the internet by various providers. Basemaps serve as a reference map on which you overlay data from layers and visualize geographic information. Rather than appearing to float in air like Sacramento is in the maps above, a basemap “grounds” the location.
We can use the basemaps provided by the package
maptiles. It offers basemaps from a number of
providers, including Esri, CARTO and Google Maps. The default is OpenStreetMap, which is a
free, open geographic database updated and maintained by a community of
volunteers via open collaboration. We can get a basemap using the
function get_tiles().
sac_basemap <- get_tiles(saccitytracts)Within get_tiles(), you specify the geographic extent of
the basemap, which can be in the form of an sf,
SpatRaster, or SpatVector object. Note that we get a
raster object in return.
class(sac_basemap)## [1] "SpatRaster"
## attr(,"package")
## [1] "terra"We can then overlay our Sacramento City tracts object on top using
the function tm_rgb()
tm_shape(sac_basemap) +
tm_rgb() +
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_layout(legend.outside = TRUE, legend.outside.position = "right")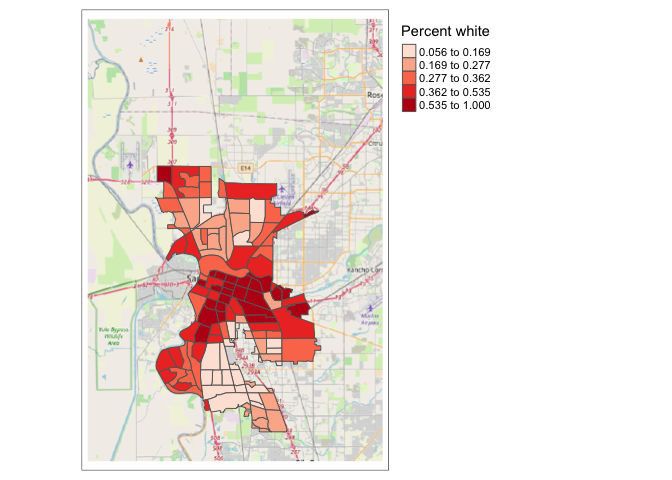
Title
Another functionality of the tm_layout() function is to
set a title for the map, and specify its position, size, etc. For
example, we can set the title, the title.size and the
title.position as in the example below. We made the font
size a bit smaller (0.8) in order not to overwhelm the map, and
positioned it in the bottom right-hand corner.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_layout(title = "Percent white Sacramento tracts", title.size = 0.8,
title.position = c("right","bottom"),
legend.outside = TRUE, legend.outside.position = "right")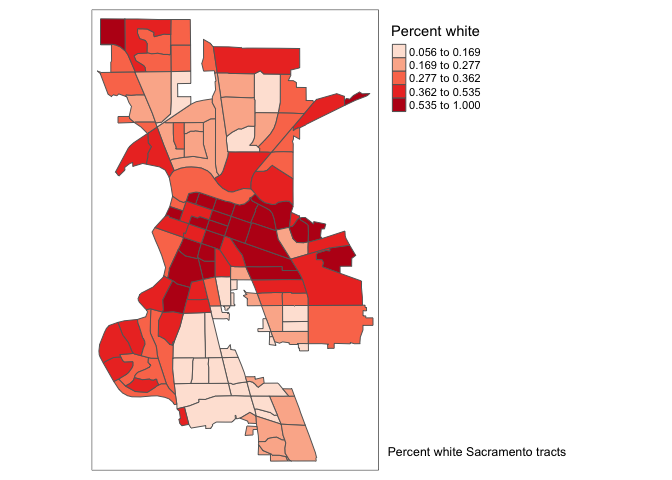
To have a title appear on top (or on the bottom) of the map, we need
to set the main.title argument of the
tm_layout() function, with the associated
main.title.position, as illustrated below (with
title.size set to 1.25 to have a larger font).
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_layout(main.title = "Percent white Sacramento tracts",
main.title.size = 1.25, main.title.position="center",
legend.outside = TRUE, legend.outside.position = "right")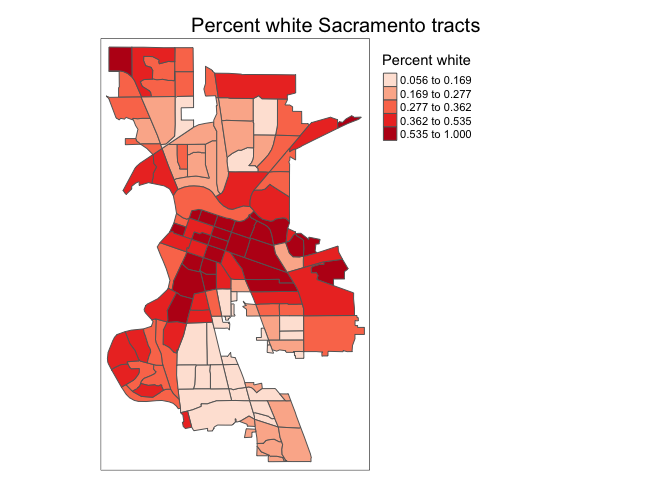
Other features
We need to add other key elements to the map. First, the scale bar,
which you can add using the function tm_scale_bar(). Scale
bars provide a visual indication of the size of features, and distance
between features, on the map.
tm_shape(saccitytracts, unit = "mi") +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_scale_bar(breaks = c(0, 1, 2), text.size = 1,
position = c("left", "bottom")) +
tm_layout(main.title = "Percent white Sacramento tracts",
main.title.size = 1.25, main.title.position="center",
legend.outside = TRUE, legend.outside.position = "right")
The argument breaks tells R the distances to break up
and end the bar. Make sure you use reasonable break points. Sacramento
is not, for example, 500 miles wide, so you should not use
c(0,10,500) (try it and see what happens. You won’t like
it). Note that the scale is in miles (were in America!). The default is
in kilometers (the rest of the world!), but you can specify the units
within tm_shape() using the argument unit.
Here, we used unit = "mi". The position =
argument locates the scale bar on the bottom left of the map.
We can also add a north arrow, which we can add using the function
tm_compass(). You can control for the type, size and
location of the arrow within this function. I place a 4-star arrow on
the bottom right of the map.
tm_shape(saccitytracts, unit = "mi") +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_scale_bar(breaks = c(0, 1, 2), text.size = 1,
position = c("left", "bottom")) +
tm_compass(type = "4star", position = c("right", "bottom")) +
tm_layout(main.title = "Percent white Sacramento tracts",
main.title.size = 1.25, main.title.position="center",
legend.outside = TRUE, legend.outside.position = "right")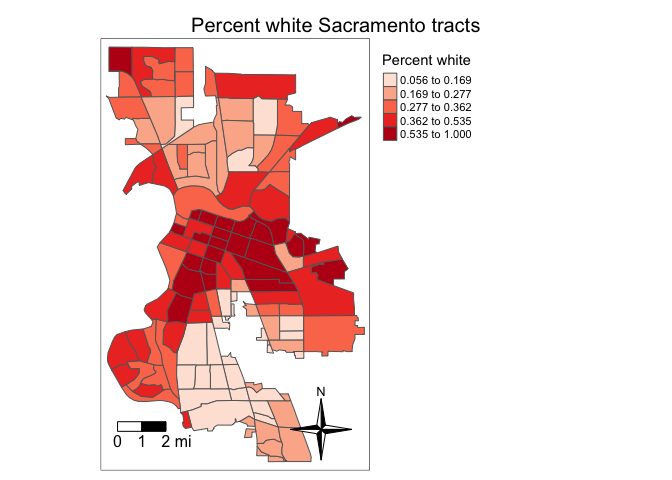
We can also eliminate the frame around the map using the argument
frame = FALSE within tm_layou().
tm_shape(saccitytracts, unit = "mi") +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_scale_bar(breaks = c(0, 1, 2), text.size = 1,
position = c("left", "bottom")) +
tm_compass(type = "4star", position = c("right", "bottom")) +
tm_layout(main.title = "Percent white Sacramento tracts",
main.title.size = 1.25, main.title.position="center",
legend.outside = TRUE, legend.outside.position = "right",
frame = FALSE)
Finally, you can add a reference or credit to your map data using the
function tm_credits(). Similar to the other map features,
we can adjust the size, positioning, etc. of the credits.
tm_shape(saccitytracts, unit = "mi") +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_scale_bar(breaks = c(0, 1, 2), text.size = 1,
position = c("left", "bottom")) +
tm_compass(type = "4star", position = c("right", "bottom")) +
tm_layout(main.title = "Percent white Sacramento tracts",
main.title.size = 1.25, main.title.position="center",
legend.outside = TRUE, legend.outside.position = "right",
frame = FALSE) +
tm_credits("Source: American Community Survey", size = 0.5,
position= c("left", "bottom"))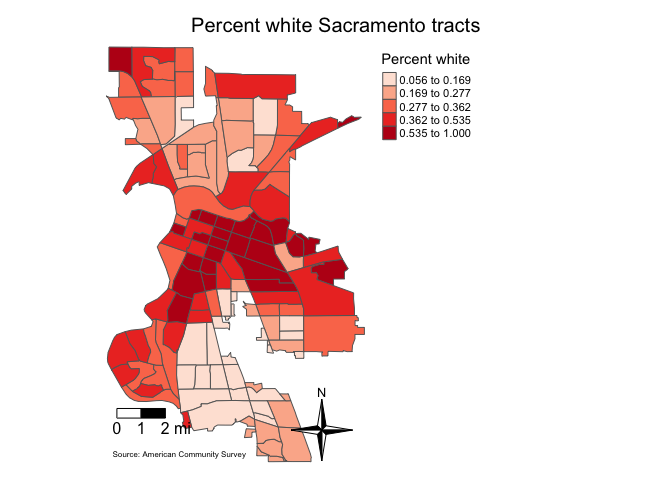
So far we’ve created static maps. That is, maps that don’t “move”. But, we’re all likely used to Google or Bing maps - maps that we can move around and zoom into. You can make interactive maps in R using the package tmap. Here is another benefit of using tmap over ggplot - the latter does not provide interactivity.
To make your tmap object interactive, use the
function tmap_mode() and specify “view”.
tmap_mode("view")Now that the interactive mode has been ‘turned on’, all maps produced
with tm_shape() will launch.
tm_shape(saccitytracts) +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
border.alpha = 0, title = "Percent white") Zoom in and out to find out where percent white is high and low. To switch back to plotting mode (static), type in
tmap_mode("plot")Behind the scenes, the package leaflet is used for generating the interactive map
Visualizing raster data
In tmap, single-band raster data are symbolized
using tm_raster(). Here, we plot the raster data set
elev we brought earlier, which represents elevation in
Luxembourg.
tm_shape(elev)+
tm_raster()+
tm_layout(legend.outside = TRUE)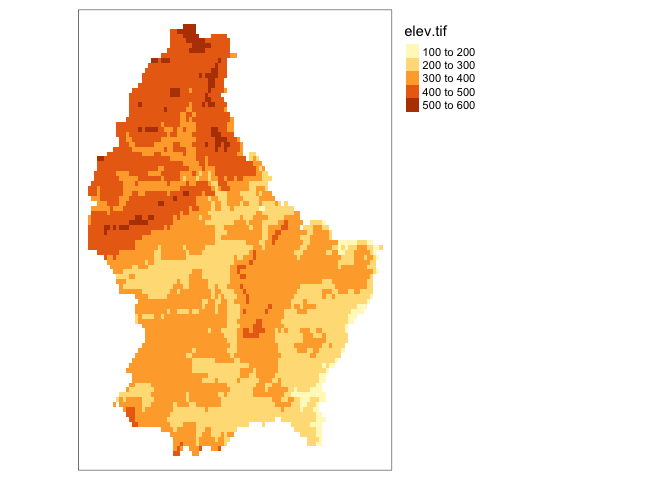
We can use all the same features discussed above for areal data with
raster data. For example, we can adjust style
tm_shape(elev)+
tm_raster(style = "quantile")+
tm_layout(legend.outside = TRUE)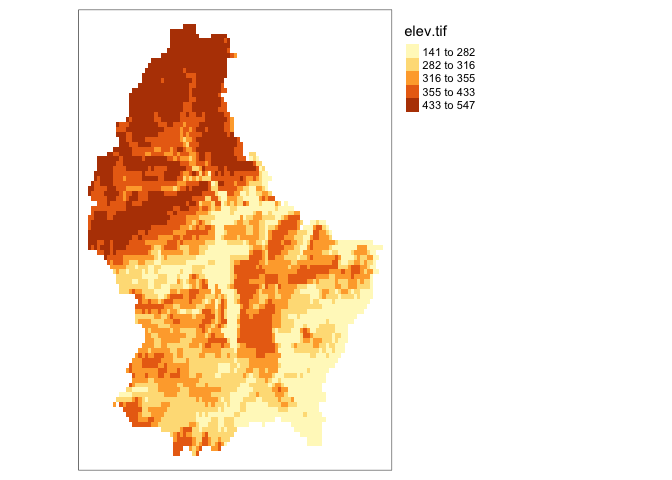
If you do not want to group colors into ranges or bins, you can use the continuous method where a color ramp is used to display the data without binning.
tm_shape(elev)+
tm_raster(style= "cont")+
tm_layout(legend.outside = TRUE)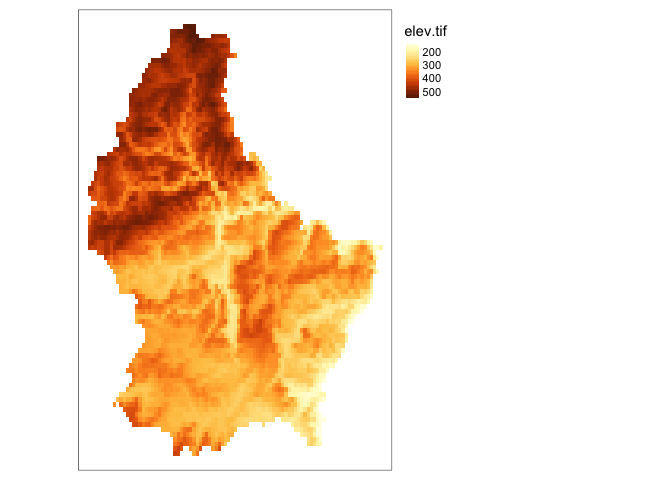
We can also adjust the color. Here, we use brewer.pal to
obtain a grey color scheme.
tm_shape(elev)+
tm_raster(style= "quantile", n=7, palette=brewer.pal("Greys", n = 7))+
tm_layout(legend.outside = TRUE)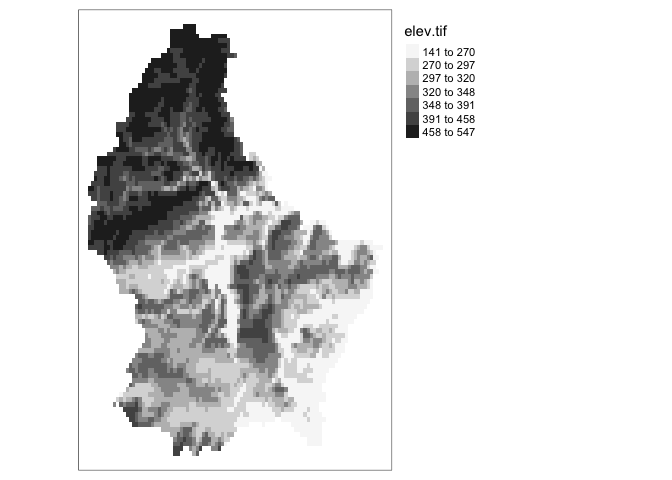
We can also visualize categorical raster data. First, lets create a raster with a categorical variable, in this case regional name.
region <- rasterize(lux, r, 'NAME_2')Next, we can map this variable by setting the style argument equal to cat.
tm_shape(region)+
tm_raster(style= "cat", title="Region")+
tm_layout(legend.outside = TRUE)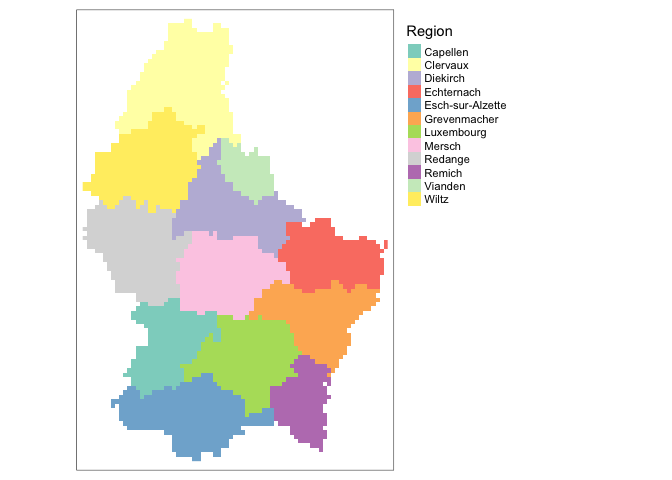
Of course, region is a bit of a trivial example of a raster categorical variable. Something like land cover type or a variable that is spatially granular or best represented as a grid is a better use of a raster categorical representation.
Dot map
OSU talks about dot maps on pages 66-71. We can create a dot or pin
map using tm_dots(). Let’s do so for EV chargers in
Sacramento. We’ll overlay it on top of Sacramento city tract
borders.
tm_shape(saccitytracts) +
tm_borders() +
tm_shape(evcharge) +
tm_dots(col = "red", title="Environmental Exposure Type") +
tm_layout(main.title = "EV Charge Stations Sacramento",
main.title.size = 1, main.title.position="center")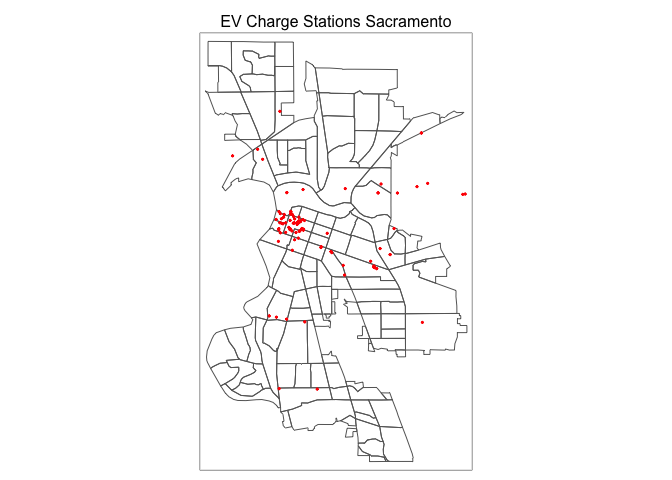
Let’s overlay it on top of polygons shaded by percent white.
tm_shape(saccitytracts, unit = "mi") +
tm_polygons(col = "pwh", style = "quantile",palette = "Reds",
title = "Percent white") +
tm_shape(evcharge) +
tm_dots(col = "black") +
tm_scale_bar(breaks = c(0, 1, 2), text.size = 1,
position = c("left", "bottom")) +
tm_compass(type = "4star", position = c("right", "bottom")) +
tm_layout(main.title = "EV Charge Stations Sacramento",
main.title.size = 1.25, main.title.position="center",
legend.outside = TRUE, legend.outside.position = "right",
frame = FALSE)
Color patch map
OSU page 72 discusses color patch maps, which is a map of polygons
based on a categorical variable. To do this, use
style = "cat". Let’s map the categorical variable
medinc, which has categories “High”, “Medium” and “Low”.
tm_shape(saccitytracts) +
tm_polygons("medinc",style="cat",palette="Paired") +
tm_layout(main.title = "Median Income Sacramento",
main.title.size = 1, main.title.position="left",
legend.outside = TRUE, legend.outside.position = "right")
Cartogram
In R, a useful implementation of different types of cartograms is
included in the package cartogram. Specifically, this
supports the area cartograms described in OSU page 78 using
cartogram_cont(). The result of these functions is a simple
features layer, which can then be mapped by means of the usual
tmap commands.
For example, we take the pwh variable to construct an area
cartogram. First, we need to add a projection
to saccitytracts. We do this by using
st_transform(), and we use the UTM
projection.
saccitytracts <-st_transform(saccitytracts,
crs = "+proj=utm +zone=10 +datum=NAD83 +ellps=GRS80") Next, we pass the layer (saccitytracts) and the variable to
the cartogram_cont function. We check that the result is an
sf object.
carto.cont <- cartogram_cont(saccitytracts,"pwh")
class(carto.cont)Which we can map. Is the cartogram a useful visual tool for a variable like percent white?
tm_shape(carto.cont) +
tm_fill("pwh") +
tm_borders()
Writing spatial data
Earlier you learned how to read in spatial data. We end this lab
guide by learning how to write spatial data. The function is
st_write().
Let’s save the Sacramento tract sf object
saccitytracts as a shapefile .shp.
st_write(saccitytracts, "Sacramento_Tract_pwhite.shp", delete_layer = TRUE)The delete_layer = TRUE argument tells R to overwrite
the file Sacramento_Tract_pwhite.shp if it already exists in
your current directory (use getwd() to check your current
working directory). You should see the files associated with
Sacramento_Tract_pwhite in your current working directory.
Answer the assignment 5 questions that are uploaded on Canvas. Submit an R Markdown and its knitted document on Canvas by the assignment due date.
Old spatial packages
If you search online for help and resources on spatial data in R, you
will find information on a number of older packages that users relied on
to process, wrangle and analyze spatial data. Before the
sf package was developed, the sp
package was used to represent and work with vector spatial data. This
package is still used to some extent, but is getting phased out by
sf. Nevertheless, if you are confronted with managing
spatial data in the sp world, check out their official
CRAN
page. The st_as_sf() function in sf
can be used to transform an sp object to an
sf object. sp as well as the
rgdal, rgeos, raster
and maptools packages are no longer maintained and will
retire.
Resources
This is not a Geographic Information Science class, so we did not touch on the many tools that sf and terra offer for manipulating and managing your spatial data. The best references for cleaning, managing and analyzing spatial data in R are Geocomputation with R (GWR) and Spatial Data Science (SDS). Both are freely available through the links provided.
For more on using ggplot() to map, check out this
and this.

This
work is licensed under a
Creative
Commons Attribution-NonCommercial 4.0 International License.
Website created and maintained by Noli Brazil