Assignment Guidelines
GEO 200CN - Quantitative Geography
GEO 200CN - Quantitative Geography
Assignments are released Monday morning as pdfs on Canvas in their respective weekly Lab and Assignment folder. They will be typically due the following Monday morning (see the course agenda in the syllabus). Assignments will typically (but not always) have two components:
- Answering non R-related conceptual questions.
- Executing data analysis tasks in R - for example, create a map or run a statistical model and interpret its results.
For (2), sometimes the questions are outside of the lab guide, while sometimes they are embedded within the lab guide.
You will use R Markdown to write up all assignments, including the non-R related questions.
R Scripts
Running R code using the console is a great place to start, but has its limitations. Each time you want to execute a set of commands, you have to re-enter them at the command line. Complex commands are potentially subject to typographical errors, necessitating that they be re-entered correctly. Repeating a set of operations requires re-entering the code stream. To give yourself more room to work, it’s a great idea to use the R script editor.
A script is simply a text file containing a set of commands and comments. The script can be saved and used later to re-execute the saved commands. The script can also be edited so you can execute a modified version of the commands.
You can open a new empty script by clicking the New File icon in the upper left of the main RStudio toolbar. This icon looks like a white square with a white plus sign in a green circle. Clicking the icon opens the New File Menu. Click the R Script menu option and the script editor will open with an empty script. Your R Studio should look similar to below (image source: R for Data Science)
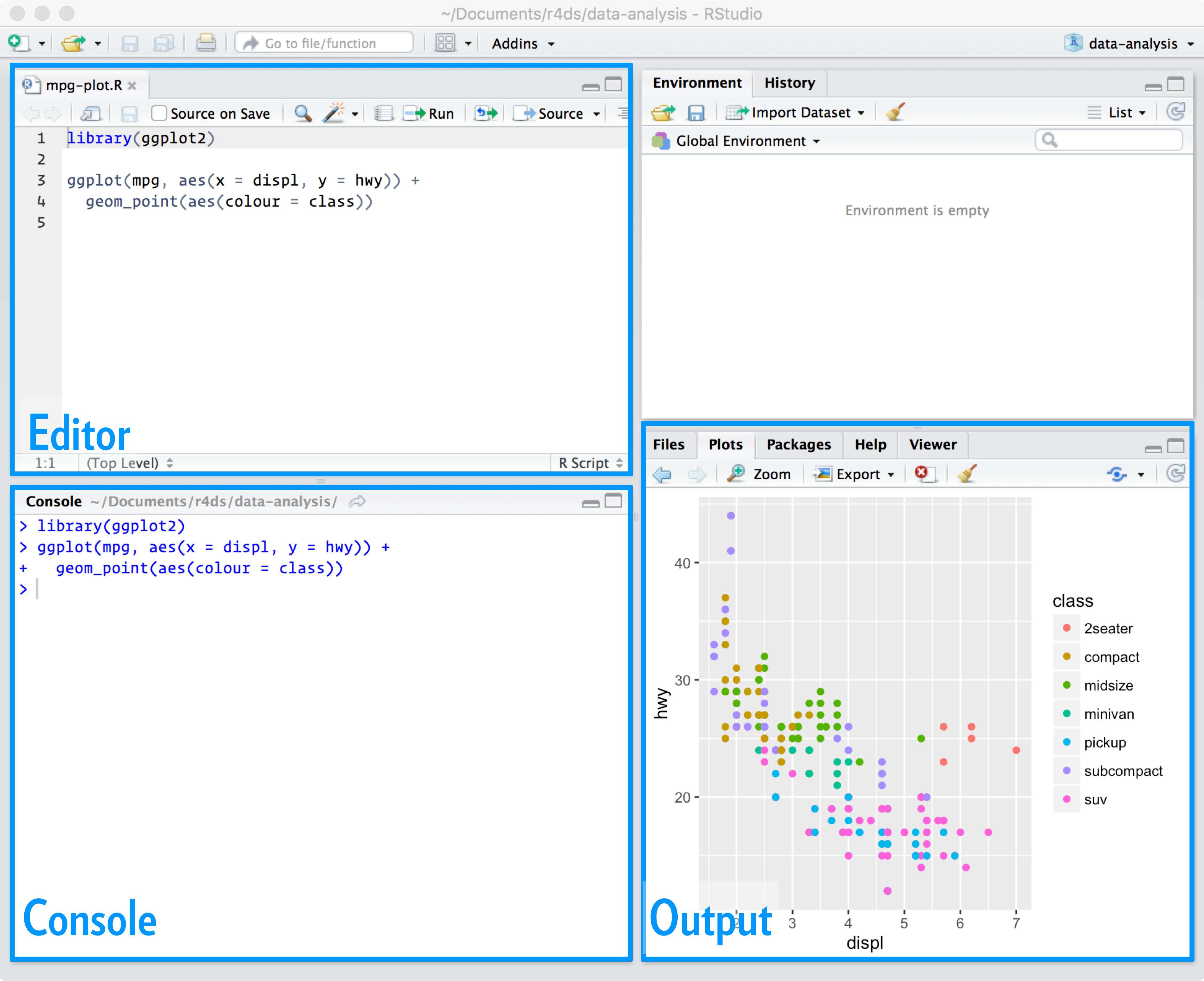
The panel that pops up is the mystery fourth window I was referring to in the Getting Started guide. Once the new script opens in the Script Editor panel, the script is ready for text entry.
R Scripts are basically text files with a .R extension that houses your code and comments. You write your code. Execute that code from your R script. Results are produced in your R console window. In this class, we won’t be using regular R Scripts for homework assignments. Instead, we’ll be using R Markdown.
R Markdown documents take script files to a new level by allowing you to mix R commands with explanatory text. Think of an R Markdown document as an R script on steroids. Your R Markdown source document is compiled into an output report evaluating the R commands in the source document to produce easily reproducible results in an aesthetically pleasing form. It combines code, results from the code, and narrative text explaining the results to produce beautiful documents and academic reports.
R Markdown
R Markdown is a simple formatting syntax for authoring html, pdf, and Microsoft Word documents in RStudio. For each R related assignment, you will upload onto Canvas two documents:
- An R Markdown document, which has an
.Rmdextension - A knitted
.html,.pdfor.docxfile
These documents will provide us an easy-to-read document to grade; more importantly, you will get to practice (1) writing scripts, (2) keeping track of the analyses you run, and (3) organizing your output in a reader-friendly manner. When you submit these documents on Canvas, do not combine them into a zipped compressed folder. They should be two separate files.
To be clear, R is a programming language. RStudio is an application. R Markdown is a markup syntax to convert R script and text into a pdf or html document. It allows for presentation-ready documents that show commands and results in a seamless flow. When you write R code and embed it in presentation documents created using R Markdown, you are forced to explicitly state the steps you took to do your research.
In RStudio, install the packages knitr and
rmarkdown using the install.packages()
command. Type the following in your RConsole window after
>
install.packages("knitr")
install.packages("rmarkdown")Once you’ve installed these packages, you don’t need to install them
any more in the future. You also do not need to load them in at
any time using library().
Creating and saving an R Markdown document
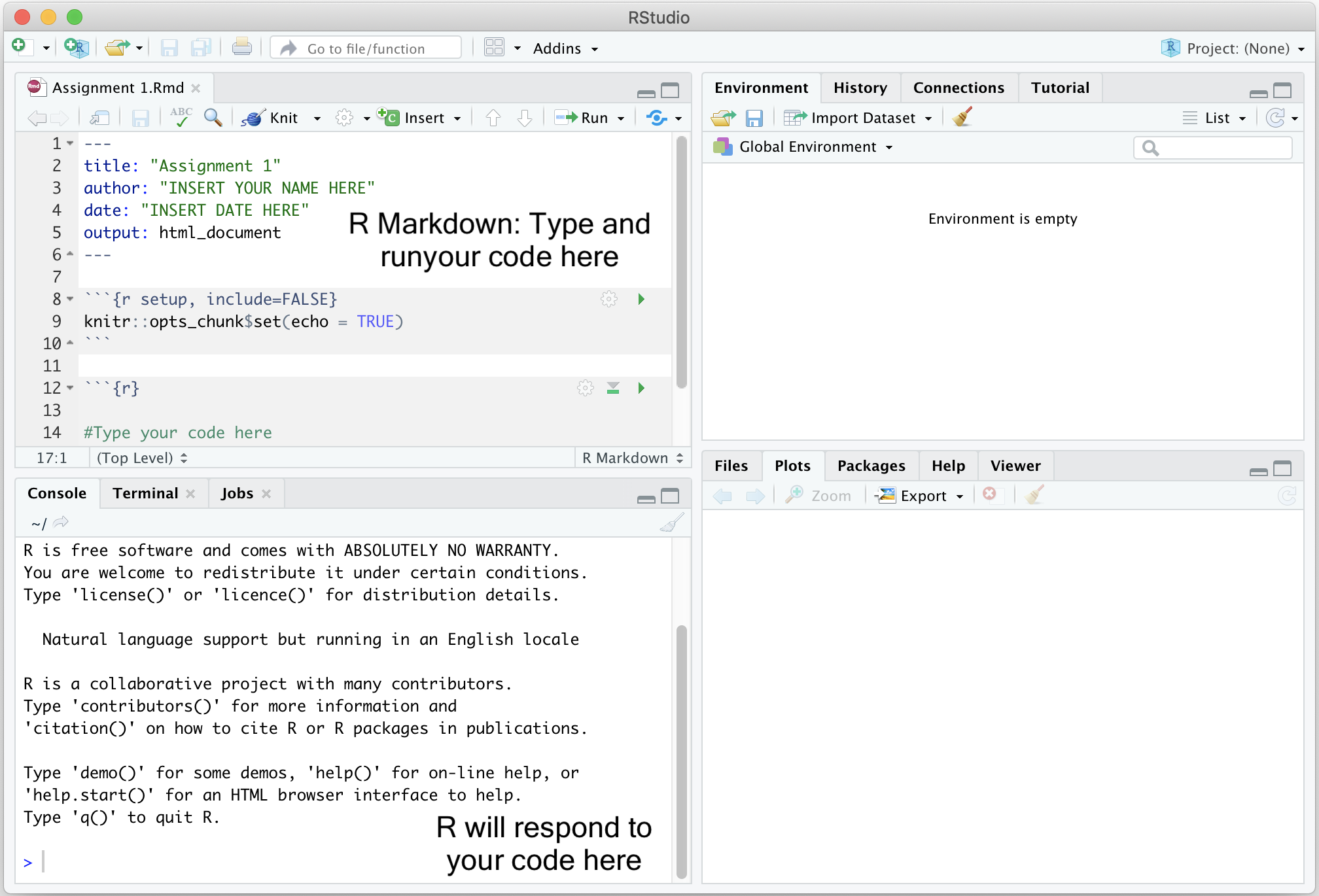
To create a new .Rmd file in RStudio, select File -> New
File -> R Markdown. A window should pop up. Type in “Assignment X”,
where X is the assignment number, next to Title and your name
next to Author. The date should be the current date. Leave
everything else alone and click OK. A new window in the top left of your
RStudio console should pop up containing your new R Markdown file.
If you are using your personal computer for this guide, it’s best to set up a clean and efficient file management structure. File management is key to preventing R programming frustration. Here are some tips.
- Set up a clear and understandable hierarchical file system for this class on your hard drive. For example, create a class folder (GEO 200CN). Within this class folder, create the folder Assignments. Within the Assignments folder, create separate folders for each Assignment (e.g. Assignment 1, Assignment 2, …). Don’t work from your Desktop. Or from the Downloads folder. Or some randomly named folder that you will have a hard time finding a month, week or day from now. You might end having a system that looks like the following (on a Mac)
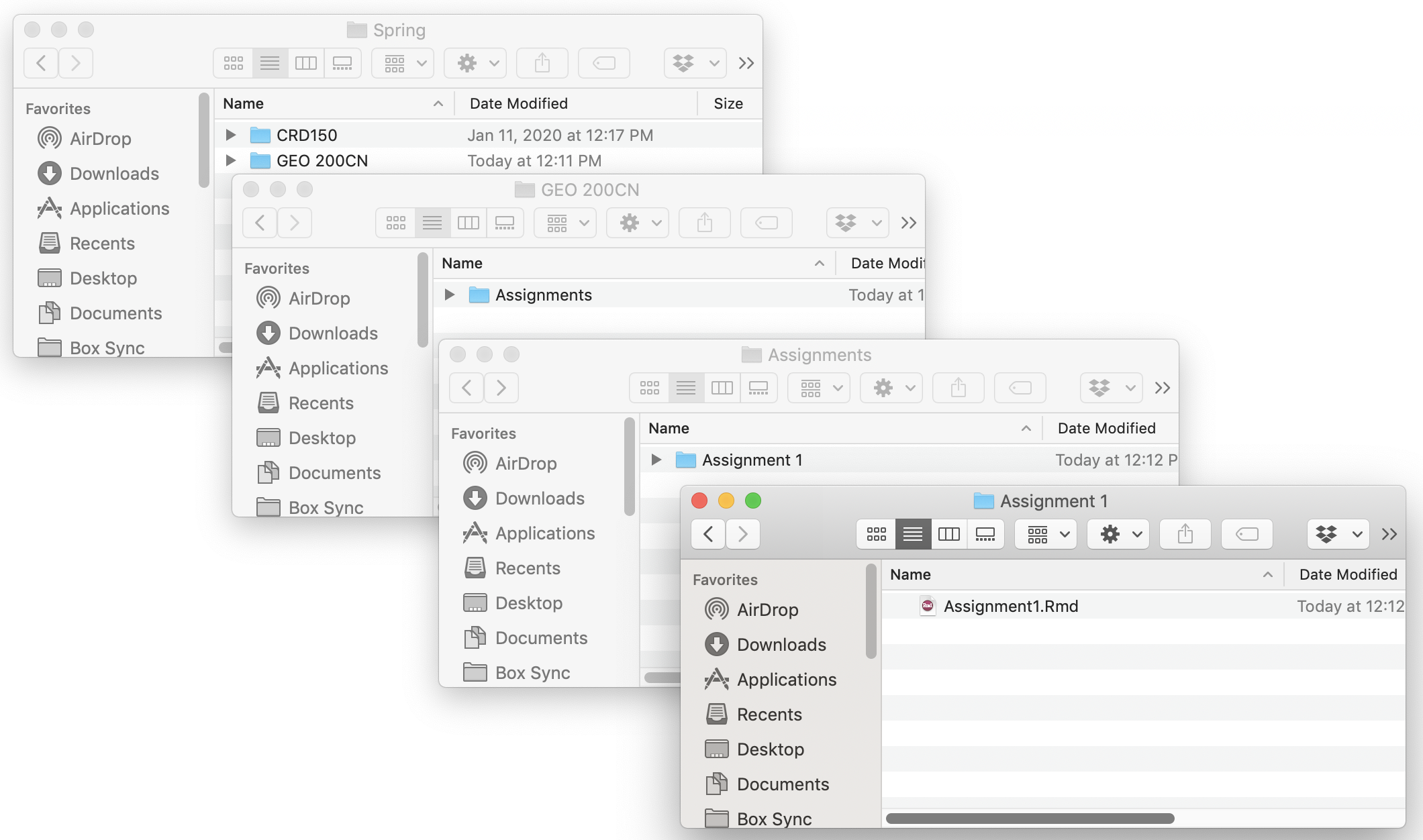
- Keep all your files that are related to a lab or assignment in one folder. That is, don’t have an Assignment 3 folder that contains data files specific to Assignment 1. Keep everything in one folder, including the R Markdown for that assignment or lab.
Using the basic guidelines above, save your R Markdown file into an appropriate folder on your hard drive by clicking on File and then Save from the RStudio menu.
Knitting an R Markdown document
In addition to the R Markdown Rmd file, you will need to submit its knitted result. Knitting puts an assignment’s main components - code, output, and text - in a nicely formatted document. You can create three types of knitted documents: html, Microsoft Word, and a pdf. I recommend knitting to an html file because it is the easiest of the three options. Go back to the YAML example I showed above. output: html_document tells R to produce an html document. If you want to knit to a pdf, you’ll need to do some extra package installing. See here.
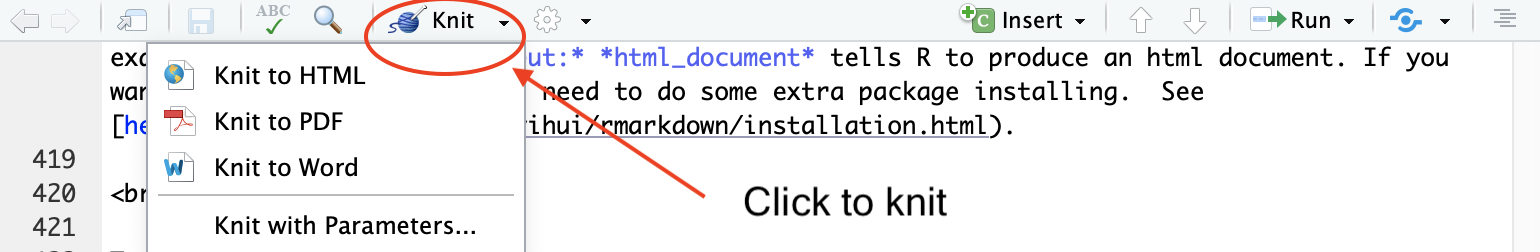
To Knit your document click  ,
which will be located at the top of the upper left R Markdown window.
Note that you can select your document type when knitting by clicking
the pull down menu next to
,
which will be located at the top of the upper left R Markdown window.
Note that you can select your document type when knitting by clicking
the pull down menu next to  and selecting your document choice (default is html).
and selecting your document choice (default is html).
When you start knitting, you will notice that a new window on the bottom left named “Render” will appear in place of the console. The window will show the progress in your knitting. If it has a problem knitting, R will stop and give you an error. Hopefully, the error statement reveals why you were not able to knit. If it is not illuminating (oh oh), you’ll need to figure out what you did wrong. See the Having problems knitting? section below.
If you encounter no errors, a preview of your knitted document will
pop up in a new window and the .html (or pdf or docx) file
will be saved in the folder where your Rmd file resides. I recommend not
waiting till the very end of an assignment to knit. When you finish one
question, knit your document to see if everything is working properly.
If it is working for that question, move on to the next question.
Let’s be clear. There are two things you’ll have to deal with: (1) Making sure the R code is working correctly to get the results you need in order to answer the question (2) Making sure the code is working correctly to knit a final document. These two issues may be related (if your R code is producing an error, R Markdown will not knit), but sometimes they are not. So, check both your R code and your knitting results often. And absolutely do not wait till the last minute to knit. Knit as often as possible.
When you’re satisfied with the end product, submit your
.Rmd document and a knitted document on Canvas.
Having problems knitting?
- A major source of error for most new R Markdown users is that they call up a data object in their R Markdown file that has not been created within the R Markdown file. Treat the R Markdown as its own separate system - even if you’ve created an object through the R Console, and you can see it sitting in your Environment window, R Markdown won’t recognize it because it was not created within the R Markdown document.
To be clear, let’s say you typed directly in the R console the following code:
myobject <- 2You see the object myobject pop up in your Environment window in the top right window. Let’s say you write in your R Markdown file the code:
```{r}
myobject*10
```You then click on  to knit. You will get an error because R will not be able to knit
because myobject was not created within the R
Markdown. Both lines of code should be in the R Markdown file
as follows:
to knit. You will get an error because R will not be able to knit
because myobject was not created within the R
Markdown. Both lines of code should be in the R Markdown file
as follows:
```{r}
myobject <- 2
myobject*10
```Once again, treat the R Markdown file as a self-contained, stand alone script. This is an important concept to understand because many students get tripped up on it when first starting out.
- Are you trying to bring in a data file that is not located in the
directory your R Markdown is pointed to? Remember, don’t scatter your
data files for an assignment across different folders. Keep them in one
folder - the folder where your R Markdown document resides. To find your
current working directory, type in
getwd()in your console and press enter. To set your working directory, type insetwd("folder")in your R Markdown and replace “folder”” with the entire path you want R to point to. You can also set your working directory by clicking on Session -> Set Working Directory -> Choose Directory from the top menu. - Do you have an
install.packages()in your R Markdown script? Take it out! You only need to do it once - and never inside an R Markdown script. - Do you have a
View()command in your R Markdown script? Take it out! Sometimes R Markdown will have problems when trying to view an R data object. - Are you using functions for a package that you need to load into R
and haven’t loaded it in your R Markdown using
library(). If so, load it in R Markdown! - If you change a piece of code somewhere in the middle because you caught an error, make sure that change is reflected throughout the code. For example, let’s say you decide to rename the object hisobject to herobject. There may be code later on in the document that uses hisobject. So, you’ll need to change every line of code in your R Markdown that uses hisobject to now use herobject.
- SPELLING, Spelling, spelling. Check your spelling. You created an
object named hisobject, but you use it later in the file as
hsobject. R will tell you that hsobject is not found.
You use the function
read_cvs()as opposed to the correct functionread_csv().
- R is case sensitive. The object Money is different from money.
- R functions are sometimes finicky about spaces. They are also
finicky about quotes. Some functions require quotes, others don’t.
- Make sure that when you use brackets, parentheses or quotes, they should start and end with one. It is very rare that you will use a left parentheses and not close it with a right parentheses.
- As I mentioned above, don’t wait till the last minute to knit. Knit after every question. Repeat it again: Knit after every question.
- The first place to check when you get a knitting error is the specific chunk or line that the error is pointing to.
- As I mentioned above, check if your R code works one chunk at a time.
- Having problem with a line of R code?
- Did you install the appropriate package?
- Did you load in the appropriate library?
- Are you using the right function?
- Did you specify all the function’s arguments correctly?
- Still having problems? Break up your code line by line or even argument by argument to find the error? For example, let’s say you have 4 lines of code that are connected together - i.e. line 4 depends on line 3, line 3 depends on line 2, and so on
line 1 code
line 2 code
line 3 code
line 4 codeIf you get an error, run line 1 first. No error? Run line 1 and 2. No error? Keep going until you find the offending line.
- If you have a Mac and you are getting an error when knitting, you may need to download the most recent version of XQuartz, which can be downloaded here
- If you’re still stuck, more than likely someone else also had a similar problem in the past. So, ask a classmate. If your classmate is also stuck, use Google and it might help you out.
Summary
The proper workflow for each assignment will be as follows
- Go through the week’s reading and lab guide and make sure you understand the material.
- Create a folder on your hard drive that is specific to the assignment (e.g. Assignment 1, Assignment 2, etc.).
- Create an R Markdown assignment file. Save it in the appropriate assignment folder on your hard drive.
- Download any data needed for the assignment into the same folder. For most assignments, I will upload the assignment data on GitHub, which you can directly link to in R, so you won’t have to download data. Some files will be located on Canvas.
- In the R Markdown document, answer the first assignment question.
- Some questions will be non R related. Answer those questions within the R Markdown as text.
- Most of the questions will ask you to run code. Some of these
questions will be outside of the lab guide.
- Other questions will be embedded in the lab guide. In these cases, you will need to run the lab guide code in your Assignment R Markdown. To be clear, some assignments, the questions will be located in the lab guide, so you will need to run the lab guide code in your Assignment R Markdown.
- Show your code in R Markdown chunks. Bottom line: Any code you used
to get a result should be in your assignment. Otherwise, you will get
points off, and for some questions, get all points off.
- Break up your code into different chunks where it makes sense. For some questions, you might include all code for a question in one single chunk. For other questions, you might break up the code into several chunks.
- Make sure your code works. Run code one chunk at a time to make sure it is working. Note that there are multiple ways to get to an answer in R. We will not grade on how efficient your code is unless stated so in the question.
- Make sure to comment your code.
- Most of the questions will ask you to explain your results. Write your explanations outside of the R code chunks. Please - please - take these interpretation questions seriously. This is a not a programming or Data Science course - you may have taken 40 hours to produce super elegant code to answer a question, but your results won’t be worth much to anyone if you can’t properly interpret them.
- After you’ve completed the first question, knit to an html, pdf or docx file. Make sure it knits properly. If it does not, examine the error, and fix the problem.
- If you’re satisfied with your code and its results for the first question, and the document properly knitted, move on to the next question. Repeat steps 5 and 6.
- Once you’ve completed all questions and successfully knitted, submit
the
.Rmdand the knitted files on Canvas before the designated due time.
Grading
In order to get full credit for R related questions, you will need to
- Show the correct statistical results for a given question (e.g. map, table, statistics).
- Show the code producing the results.
- Annotate your code by placing explanatory text after
#in an R chunk. These annotations will allow us to know that you know what the code is doing. - Provide correct written answers.
Any response requiring a data analysis task must be supported by code you generate to produce your result. Just examining your various objects in the “Environment” section of R Studio is insufficient—you must use scripted commands. Because there are typically multiple ways to get an answer, we will not grade you on the efficiency of your code. You can ask for help from the TA and work with other students. However, you must submit your own assignments.
You must include in your own words comments that explain each line of code in your R Markdown. It does not have to be extensive, but should be concise and clear enough to let us know that you understand what the code is doing. You should indicate not just the what, but also the why. For example
```{r q1chunk1}
#keeps the variables var1 and var2 because these are the only variables I need
#for the analysis.
object <- object %>%
select(var1, var2)
```Comments are important for demonstrating that you understand what your code is doing. Furthermore, by writing in basic English what each piece of code is doing and why you are doing it, you will be able to strengthen your understanding of how R works. Furthermore, comments will help clear up any confusion when future you goes back to your code.
We will be lenient in our grading early in the quarter as you learn R. However, grading will get progressively stringent, particularly on aspects of your submission that we’ve already covered in a prior week.
Also note: all code used to produce your results must be shown in
your html or pdf file (e.g., do not use echo=FALSE or
include=FALSE as options anywhere). Other pieces of
advice
- Answers should be correct. This is not to say that we will not take into consideration how much of your code is correct. But, if you do get the wrong answer we will take at least something off.
- We will not grade on how efficient your code is unless stated so in the question. We will never take points off for accomplishing things differently than expected as long as it produces the correct result. However, if the answer is incorrect and your code indicates little effort (e.g. blank or a few lines of code that yield nothing) or is completely incomprehensible (e.g. 20 lines of mish mash), expect few to no points.
- Interpretation of results is very important. This is not a
programming class. You might have the fanciest code in the world, but it
will not matter much if you do not understand what the results are
telling you.
- You must submit both an Rmd and knitted file. If you don’t, we will take significant points off.
Other things to know
- Please spell-check your assignment before handing it in (Edit -> Check Spelling from the top menu bar).
- The default editor settings are to insert matching parentheses and quotes; if you find that this intrudes on your workflow, you can disable it via Tools -> Global Options -> Code then uncheck Insert matching parens/quotes.
- We expect you to work together. Two heads are better than one, and five heads are better than two
- We expect you to ask for help if you get stuck (and you will be stuck a lot). We can’t read minds, so if you don’t tell us that you are confused, we won’t know.
- You should try and solve problems before you ask for help.
- Trying includes an Internet search (a crucial skill!)
- All homework should be submitted on Canvas.
Getting R Markdown Help
Whenever you are editing R Markdown documents in RStudio, you can display an R Markdown cheat sheet by going to Help -> Cheatsheets -> R Markdown Cheat Sheet. A basic introduction to R Markdown can also be found in Chapter 27 in R for Data Science. R Studio has a quick introduction to R Markdown. We’ve put together some additional R tips here in the likely case that you get stuck. And, of course, there is the Google machine. Use it to get help.

This
work is licensed under a
Creative
Commons Attribution-NonCommercial 4.0 International License.
Website created and maintained by Noli Brazil
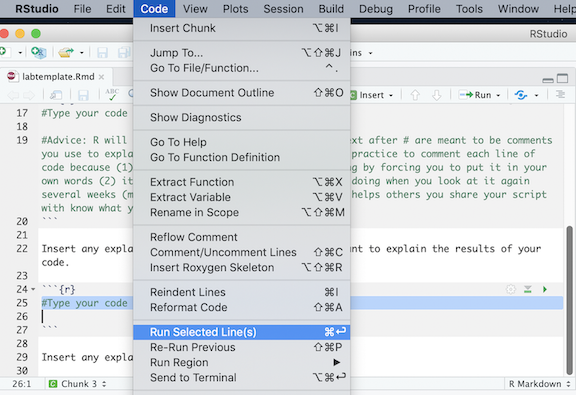
 located at the top of the R Markdown window and select Run Current
Chunk.
located at the top of the R Markdown window and select Run Current
Chunk. located on the right corner of the chunk. See Figure below.
located on the right corner of the chunk. See Figure below.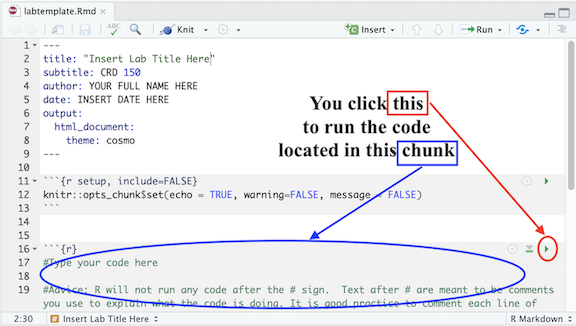
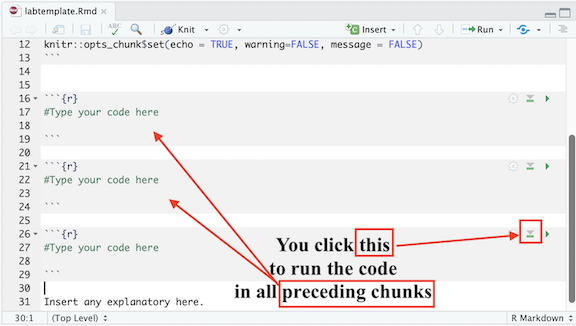
 will run all previous R chunks. See Figure below.
will run all previous R chunks. See Figure below.